Monitoring progress in translational bioinformatics
It is with great enthusiasm that the PLOS Computational Biology Education Editors present this invited blog post from Russ Altman, in what we hope will be a yearly feature for PLOS Biologue. It is a recapture of his annual review on translational bioinformatics, a topic very close to our interests and the focus of the first PLOS online book, part of the PLOS Computational Biology Education section. We hope you enjoy this blog post, and find this a useful and convenient way for us to share this information and perspective with you.
Joanne Fox and B.F. Francis Ouellette, Education Editors, PLOS Computational Biology
Each year the American Medical Informatics Association (AMIA) holds a meeting on Translational Bioinformatics as part of its “Joint Summits on Translational Science”, along with a meeting on Clinical Research Informatics. For the last several years, I have been invited to present a “Year in Review” for Translational Bioinformatics in which I summarize notable papers within translational bioinformatics during the previous 12-14 months. The meeting happens in March, so the summary usually covers the preceding calendar year plus some extra weeks.
This activity is both rewarding and quite stressful, as there is always a large body of work to review, and I would like to do a good job highlighting the work that is novel and exciting, while not just automatically choosing papers published in high impact journals. I have developed some rules to make this manageable. Primarily, I have a fairly strict definition of translational bioinformatics: the candidate papers should present a novel methodological approach to combining clinical entities (patients, diseases, drugs, signs, symptoms) and molecular/cellular entities (genes, proteins, RNA, DNA, small molecules, pathways, networks). After all, translational science is the study of how we move discoveries from bench to bedside, so translational bioinformatics should be informatics work that does the same. If there are no clinical entities, then the paper is not eligible. If there are no biological entities, then the paper is not eligible. I prefer novel informatics methodological content, but will allow papers that use off-the-shelf methods to do something really remarkable.
I try to track papers all year, but honestly have to do a lot of reviewing in the few weeks before the talk. The main sources of papers are two: (1) the recommendations of colleagues from whom I request nominations around January (self-nominations are OK, but nominating others is particularly valued); and (2) a set of somewhat ad hoc PubMED searches seeking papers that combine clinical/biological/informatics concepts. It is also fine for anyone to send me nominations throughout the year by email (russ.altman[at]stanford.edu) or even twitter (@rbaltman) and I have a system for tracking them. I then review several hundred articles, first at the title/abstract level (mostly to triage the ineligible ones), and then more deeply to find the contributions that most excite me. I do as many passes through the list as needed to reduce it to a length that allows me to present it in one hour. This is usually about 35 papers with an additional 10-15 “shout outs”, which I mention only by title.
After I have the final list of papers, I assemble them into 5-8 ad hoc groupings that provide some structure for the talk. I then create a slide deck with a rigid format: every paper is summarized by a single slide with title, first author, journal and PubMED ID, and then my bulleted summary of:
- Goal: what are they trying to do?
- Method: how did they approach the problem methodologically?
- Result: what did they find?
- Conclusion: what should we take away from the paper?
I stress that the “conclusion” is my own conclusion, not necessarily the conclusion of the authors, and functions as my pulpit to justify why I chose the paper. The next 1-2 slides are always key graphics from the paper that illustrate or summarize what impressed me about it.
So what papers did I highlight from Jan 2013 to March 2014? Well, the complete slide deck is available at my blog “Building Confidence”, as are all the slide decks from 2008 to 2014 (see QR code in the final slide of this blog). Just to provide some comparison and baseline for this year, the topics last year (January 2012 to March 2013) were:
- Omics medicine
- Cool methods
- Cancer
- Drugs
- Delivery (of healthcare)
And the topics this year were:
- Controversies
This was a new category this year, and I used i t to highlight two non-publications. I lead off with the FDA letter to the direct-to-consumer genetic testing company, 23andme, which is mandatory reading for anyone interested in translational genomics. Next, I highlighted the blog of colleague Lior Pachter, as he engaged in an entertaining (and informative) polemic about network science applied to cell biology.
t to highlight two non-publications. I lead off with the FDA letter to the direct-to-consumer genetic testing company, 23andme, which is mandatory reading for anyone interested in translational genomics. Next, I highlighted the blog of colleague Lior Pachter, as he engaged in an entertaining (and informative) polemic about network science applied to cell biology.
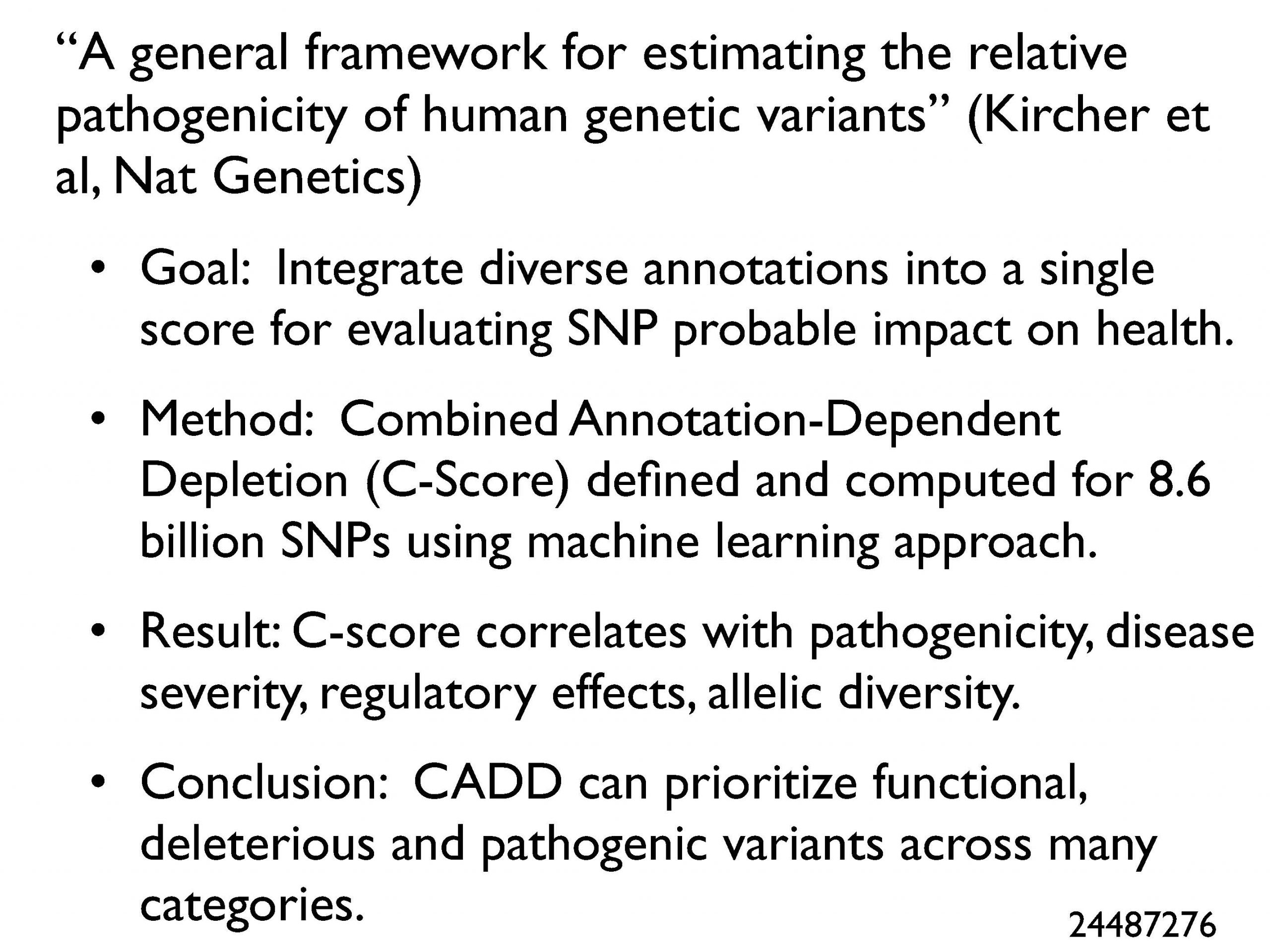
Here I highlight informatics papers pushing the agenda of clinical genomics. This included the much-awaited results of the warfarin dosing pharmacogenomics vs. clinical algorithm trials, which were split, and an analysis of the ubiquity of pharmacogenomic variants in the general population. Another good paper, shown here, by Kircher et al., introduces the Combined Annotation-Dependent Depletion (CADD) score for evaluating the probable impact of a SNP on health.
- Drugs
There are always many good papers about using informatics to learn things about drugs, including side effects and new uses. This category had several excellent papers. One paper reported the manual curation of 88,000 scientific articles, mined for drug-disease and drug-phenotype interactions. That was impressive because of the scale, if nothing else. Another favorite  was a paper showing how publicly-available gene expression data suggested that an antidepressant may be a useful adjuvant drug in lung cancer—and it’s now in clinical trials!
was a paper showing how publicly-available gene expression data suggested that an antidepressant may be a useful adjuvant drug in lung cancer—and it’s now in clinical trials!
- Genetic basis of disease
These are all informatics papers showing methods for inferring new things about the 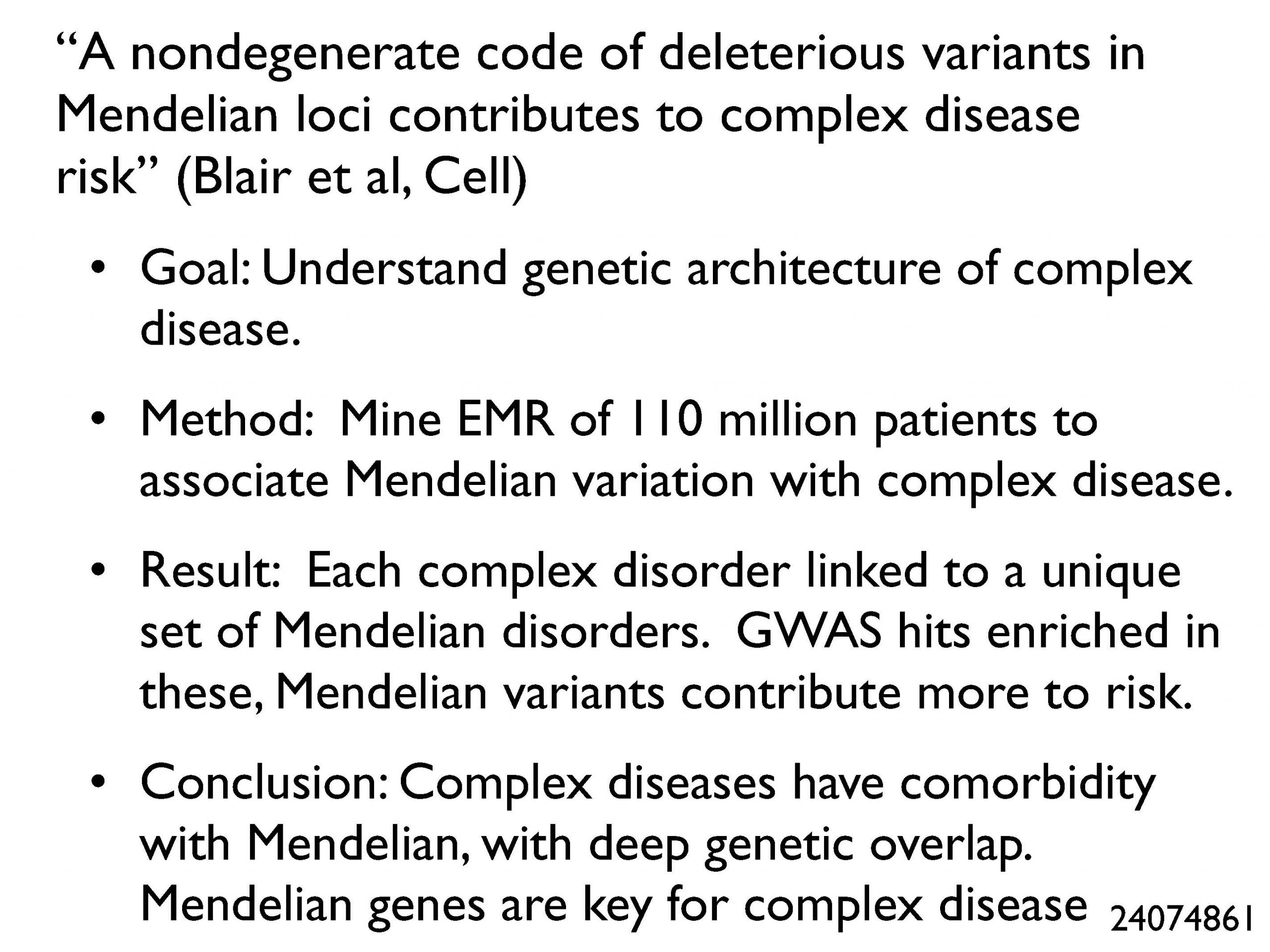 underlying mechanisms and genetics of diseases. My favorite in this category was a paper showing the many complex diseases that co-occur with Mendelian diseases, and suggesting deep genetic overlaps—as if there is a “code” in which complex diseases result from less-severe mutations in the same genes that are associated with Mendelian diseases. The implications of this work for how we interpret genome-wide association studies and think about complex disease are significant.
underlying mechanisms and genetics of diseases. My favorite in this category was a paper showing the many complex diseases that co-occur with Mendelian diseases, and suggesting deep genetic overlaps—as if there is a “code” in which complex diseases result from less-severe mutations in the same genes that are associated with Mendelian diseases. The implications of this work for how we interpret genome-wide association studies and think about complex disease are significant.
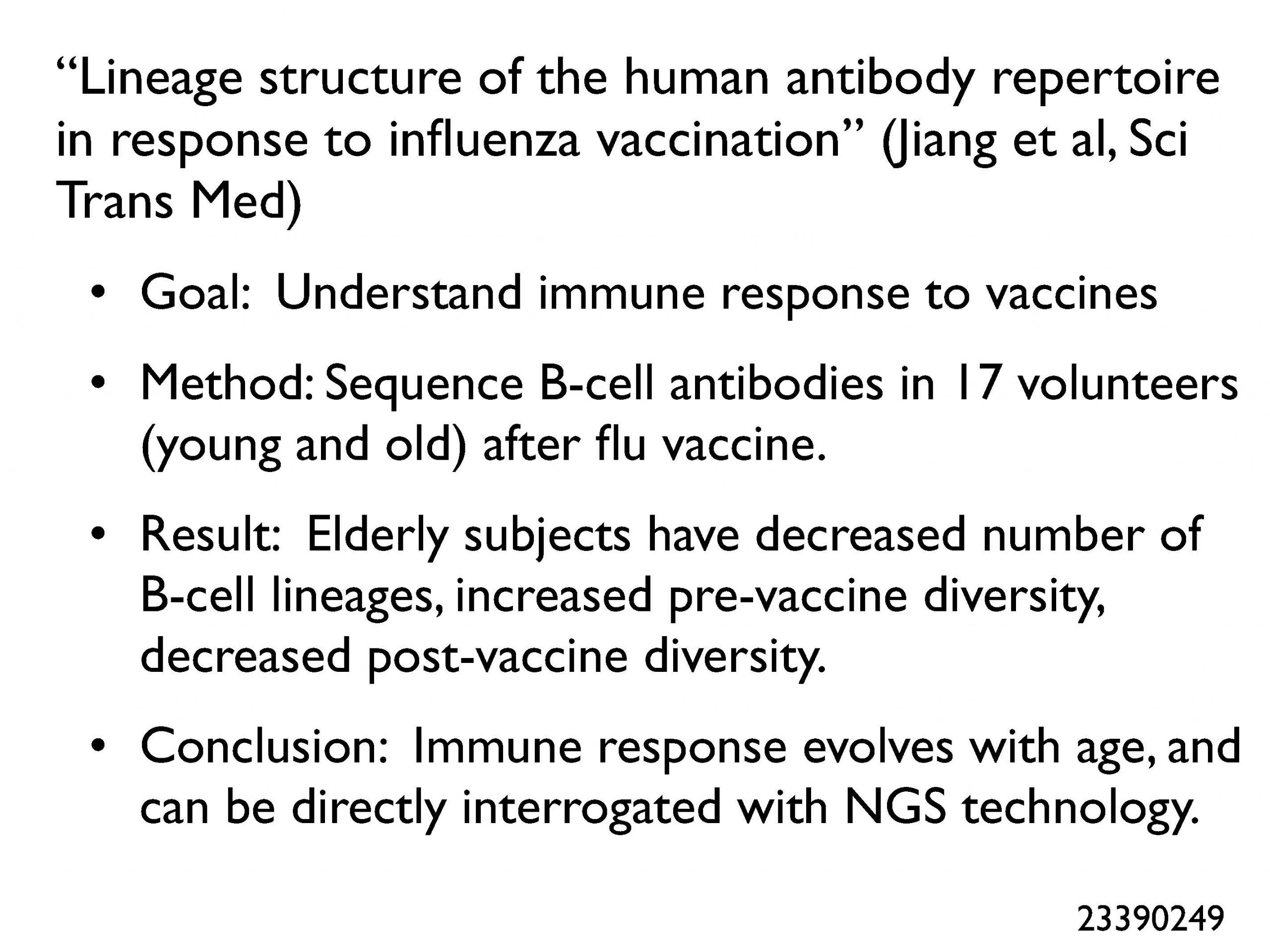
The interesting aspect of this topic is that we get a preview of the future—emerging work in the roles of long non-coding RNA, immune diversity, metabolomics, and the microbiome in disease. One exciting paper looked at the DNA sequences of B-cell antibodies and created lineage for 17 volunteers, comparing young to old in the pre- and post-vaccine state. The future is now.
- Mice. Can’t live with ‘em, can’t live without ‘em.
 One of my hard rules (usually) is that the talk will only focus on human studies and disease. This year I made an exception because of several relevant papers in mice. One praised the ability of mouse knockouts to discover/model new drugs. My favorite, however, showed that gene expression measurements on mice after burn, trauma and endotoxemia—all inflammatory conditions—not only do not correlate with similar measurements made in humans, but do not correlate with each other!
One of my hard rules (usually) is that the talk will only focus on human studies and disease. This year I made an exception because of several relevant papers in mice. One praised the ability of mouse knockouts to discover/model new drugs. My favorite, however, showed that gene expression measurements on mice after burn, trauma and endotoxemia—all inflammatory conditions—not only do not correlate with similar measurements made in humans, but do not correlate with each other!
This was a fun category that included a paper about why some publications achieve high impact, based on citation analysis. Of course, my favorite is the PLOS Computational Biology decision to publish a textbook of translational bioinformatics as a series of individual chapters. A spectacularly good idea, and a credit to PLOS.
- Odds and ends
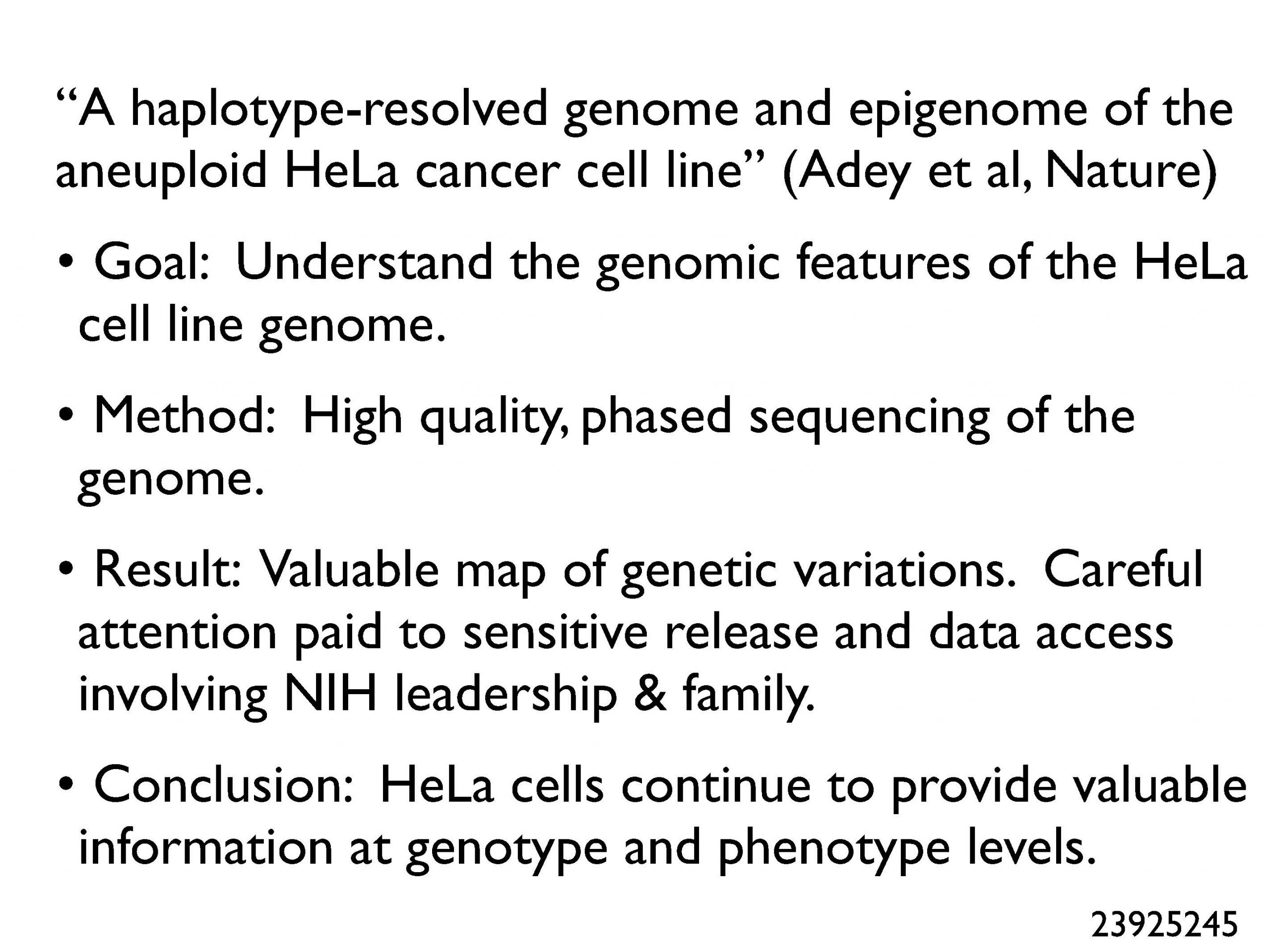 Many years require a final bucket to include important papers that don’t fit into neat categories. This year included such a category, which featured papers on social networks for tracking infections in a hospital, and for global tracking of disease based on airplanes as efficient infection-dissemination vehicles. A very important set of papers this year were those reporting the genome sequence of the HeLa cell. We all owe a debt to Henrietta Lacks for providing such a valuable biomedical research resource (under very non-optimal conditions), and these sequences help us interpret the results of HeLa experiments more precisely and with knowledge of the genome alterations underlying the cell line.
Many years require a final bucket to include important papers that don’t fit into neat categories. This year included such a category, which featured papers on social networks for tracking infections in a hospital, and for global tracking of disease based on airplanes as efficient infection-dissemination vehicles. A very important set of papers this year were those reporting the genome sequence of the HeLa cell. We all owe a debt to Henrietta Lacks for providing such a valuable biomedical research resource (under very non-optimal conditions), and these sequences help us interpret the results of HeLa experiments more precisely and with knowledge of the genome alterations underlying the cell line.
Each talk ends with a “crystal ball” section where I speculate what we may see in the coming year, and do a scorecard for my predictions the previous year. As a wise person once said, predictions are very hard, especially about the future. This year’s predictions are:
- We will see more emphasis on non-European descent populations for discovery of disease associations;
- There will be a crowd-based discovery in translational bioinformatics;
- Methods will emerge for recommending treatment for cancer based on genome/transcriptome;
- There will be more “trained systems” (like IBM Watson) applications in the field;
- There will be drug repurposing methods that suggest more than one drug used synergistically;
- There will be more cost-effectiveness evidence for genomic medicine;
- Powerful methods focused on linking genes, targets and drug response will emerge.
That’s the summary of this year’s talk. I have been invited to repeat the talk next year, and hope to see you there.
By Russ Altman, PLOS Computational Biology Editorial Advisor.




[…] Monitoring progress in translational bioinformatics review by @rbaltman https://blogs.staging.plos.org/biologue/2014/11/13/monitoring-progress-translational-bioinformatics/ […]
[…] Monitoring progress in translational bioinformatics – It is a recapture of his annual review on translational bioinformatics … that most excite me. I do as many passes through the list as needed to reduce it to a length that allows me to present it in one hour. This is usually about 35 papers with an … […]