PLOS Biology is at ASM Microbe 2016!
This year’s American Society of Microbiology conference, ASM Microbe 2016, kicks off on June 16th, 2016 in Boston, Massachusetts, USA. PLOS Biology and the rest of the PLOS family will be in attendance and are excited to meet with the community! Come visit our booth, #722, in the main exhibitor hall of the Boston Convention and Exhibition Center for some PLOS swag and information on publishing your research in our excellent journals. We will be hosting “Meet the Editors” sessions each day, Friday the 17th through Monday the 20th, with Lauren Richardson, Associate Editor at PLOS Biology, and Meghan Byrne, Senior Editor at PLOS ONE. Hope to see you there!
PLOS Biology publishes great microbiology papers. Here are some of the editors’ favorites from recent months:
Microbial interactions between kingdoms are responsible for significant microbiome variation on the surface of plants. In this study, the authors find that highly connected microbes are most important, amplifying abiotic and host factors to cause large perturbations in the structure of microbial communities (see featured image at top). Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, et al. (2016) Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol 14(1): e1002352. http://dx.doi.org/10.1371/journal.pbio.1002352
Cellulose deconstruction helps shape the global carbon cycle; this study shows that high cellulolytic ability evolved in select lineages of the bacterial genus Streptomyces through key changes in gene content and transcriptional regulation. Book AJ, Lewin GR, McDonald BR, Takasuka TE, Wendt-Pienkowski E, et al. (2016) Evolution of High Cellulolytic Activity in Symbiotic Streptomyces through Selection of Expanded Gene Content and Coordinated Gene Expression. PLoS Biol 14(6): e1002475. http://dx.doi.org/10.1371/journal.pbio.1002475
The accumulation of multiple, seemingly redundant, bacterial quorum-sensing systems is promoted by facultative cheating behavior, this study shows; the strain with multiple systems cheats its single quorum-sensing system ancestor as a minority but returns to cooperation when in the majority. Even-Tov E, Omer Bendori S, Valastyan J, Ke X, Pollak S, et al. (2016) Social Evolution Selects for Redundancy in Bacterial Quorum Sensing. PLoS Biol 14(2): e1002386. http://dx.doi.org/10.1371/journal.pbio.1002386
This study describes the structure of a novel phosphotransacylase enzyme that facilitates the recycling of the essential cofactor acetyl-CoA within a bacterial organelle and discusses the properties of the enzyme’s active site and how it is packaged into the organelle. Erbilgin O, Sutter M, Kerfeld CA (2016) The Structural Basis of Coenzyme A Recycling in a Bacterial Organelle. PLoS Biol 14(3): e1002399. http://dx.doi.org/10.1371/journal.pbio.1002399
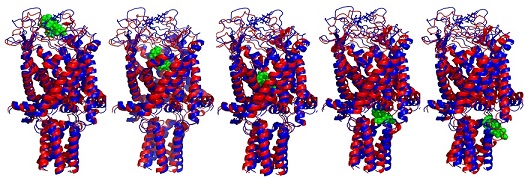
In this paper, the authors reveal that the antibiotic dihydrostreptomycin binds to a specific site on the bacterial mechanosensitive channel MscL, opens the pore, and appears to pass through to access the cytoplasm of the cell (see image above). Wray R, Iscla I, Gao Y, Li H, Wang J, et al. (2016) Dihydrostreptomycin Directly Binds to, Modulates, and Passes through the MscL Channel Pore. PLoS Biol 14(6): e1002473. http://dx.doi.org/10.1371/journal.pbio.1002473
Altruistic host bacteria can preferentially enhance the horizontal transfer of beneficial plasmids (such as those conferring antibiotic resistance or virulence) to others of their kind, as shown in this paper. Dimitriu T, Misevic D, Lotton C, Brown SP, Lindner AB, et al. (2016) Indirect Fitness Benefits Enable the Spread of Host Genes Promoting Costly Transfer of Beneficial Plasmids. PLoS Biol 14(6): e1002478. http://dx.doi.org/10.1371/journal.pbio.1002478
This study uses computational metagenomics and molecular experimentation to massively expand the known genomic and ecological diversity of RNA bacteriophages, identifying novel tropisms and genes. Krishnamurthy SR, Janowski AB, Zhao G, Barouch D, Wang D (2016) Hyperexpansion of RNA Bacteriophage Diversity. PLoS Biol 14(3): e1002409. http://dx.doi.org/10.1371/journal.pbio.1002409

Here, the authors use transposon sequencing to enable the recovery of virtually all previously characterized genes required for the differentiation of the bacterium Bacillus subtilis into a dormant spore and identifies 24 new ones. Meeske AJ, Rodrigues CDA, Brady J, Lim HC, Bernhardt TG, et al. (2016) High-Throughput Genetic Screens Identify a Large and Diverse Collection of New Sporulation Genes in Bacillus subtilis. PLoS Biol 14(1): e1002341. http://dx.doi.org/10.1371/journal.pbio.1002341
Featured Image (top): Fungal and bacterial plant symbionts (false colored yellow and green, respectively) grow in close proximity on an Arabidopsis thaliana leaf surface (yellow-red stomata). Image credit: Jonas Ruhe.
