Bad Identifiers are the Potholes of the Information Superhighway: Take-Home Lessons for Researchers
By Julie McMurry, Lilly Winfree, Melissa Haendel
This is a guest post by three of the authors of a recent PLOS Biology article, discussed here. All three authors are at the Department of Medical Informatics and Epidemiology and OHSU Library, Oregon Health & Science University, Portland, Oregon, USA.
Identifiers are (actually) important
Have you ever been overwhelmed by the sheer number of different names or symbols that point to a single gene? In today’s connected world we can easily solve this problem with the concept of an identifier – one that travels well around the Web and can include associated information such as symbols, synonyms, and the history of the nomenclature (such as when two genes were “merged” into one gene record). However, the need for best practices in the minting, use of, and referencing of identifiers isn’t limited to genes; referencing diverse types and sources of data provides a vital connection between data producers with data consumers. This connection is not an end in itself, but rather a means to improve interoperability, reproducibility, attribution, impact assessment, and knowledge synthesis — none of which is possible without identifier persistence: the ability of an identifier to continue to reliably reference the same entity. The bioinformatics world has long realized the importance of identifiers as the glue that holds scientific inquiry together both for humans and for machines. In the report published in PLOS Biology, we present a community manifesto of best practices for design, provision, and reuse of persistent identifiers to maximize utility and impact of life science data.
Identifiers are easy to get wrong
One of the most vexing issues is that there is no uniform way to represent the same identifier for the same biological entity in different contexts. This greatly complicates the tasks of how to resolve, reference, discover, cite or mention identifiers, and authors often do not recognize the importance of doing so. “Desultory referencing” has been most closely criticized for genes[1] but similarly affects references to all kinds of entities.
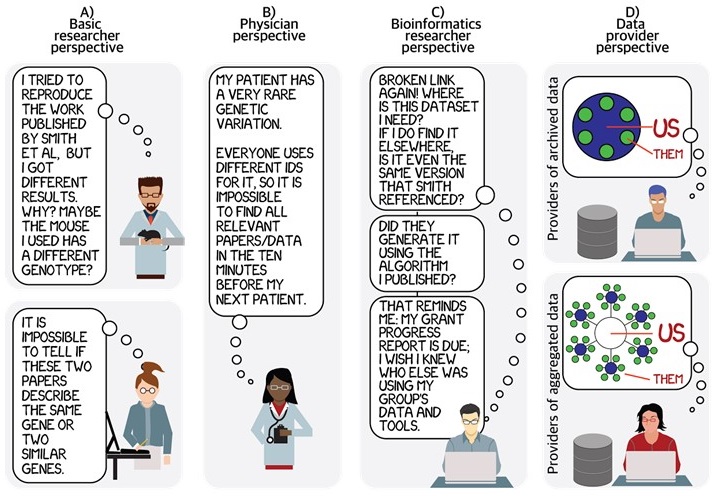
The confusion with identifiers often starts with the basics, including what the “identifier” even is. We routinely observe that “stuff falls in and out” of identifiers: all kinds of characters get prepended, subtracted, or altered when identifiers are referenced outside–and sometimes even inside–their database of origin. Identifier practices have major implications for how science gets done: if one is attempting to do a comprehensive metastudy, one may want to know all papers having to do with a specific gene, or all uses of a particular dataset, or all outputs resulting from a specific grant; all of these questions require diverse systems to work together much better than they do now. It is not without special irony that our paper on community-based identifiers was done by authors who (taken together) were supported by small portions of 19 different grants. Links to these 19 grants were documented only two years ago; however, 11 links of the 19 are now dead and had to be re-curated by hand.
Standards make the world go round. People make standards go round.
Take for example, your lighting needs. If you are an individual who wishes to read a book at night, a simple candle could suffice. A single house can subsist off-grid. However, to support the energy needs of an entire region, you’ll need a sophisticated grid and a whole lot of standards. While there is no single provider of electricity in the world, the number is finite and there exist ways to overcome the differences where needed.
Similarly, biomedical research requires increasingly sophisticated infrastructure that “just works” at global scale. Identifiers are part of this infrastructure and as such, identifier engineering faces the same challenges that most other infrastructure does: it is only ‘noticed’ when things go terribly wrong.
As with electrical work, the questions of how to create and maintain identifiers only need to be understood by those that build and maintain databases. However, everyone needs to know how electricity should be used, and this is where convention is important. Take electric devices for instance; even though there is endless variety, “1” always refers to “on” and “0” always refers to “off”. Our paper seeks to clearly define the anatomy of a persistent identifier and the practices that undergird persistence; we also make recommendations about what kinds of identifiers one can use use in specific contexts, such as when authoring a manuscript or sharing data.
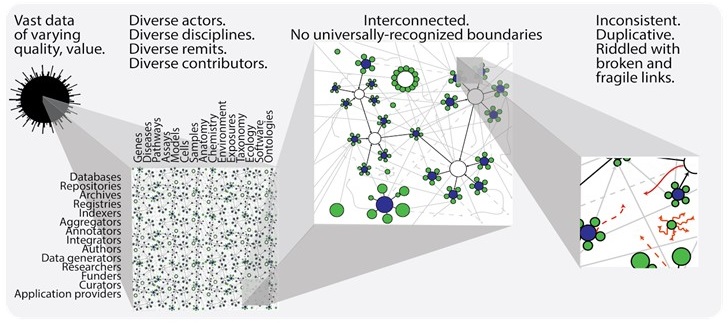
The Identifier landscape can be thought of in terms of hubs-and-spokes, however, individual communities often forget how interconnected these hubs and spokes truly are, and how fragile many of them are, leading to confusion and data errors that only get worse with time. Data is most useful when it is broken down into components and incorporated in different ways, by different users, for different purposes. However, if one piece of the identifiers landscape fails, the damage can propagate to databases/papers/applications that reference it–whether directly or indirectly.
Cite with care
When the basic framework of prefix:local_id are established, it becomes much easier to form a more coherent picture of the data across the various places it is used. For instance, if you take the identifier taxon:9606; a biomedical researcher would take this to mean human, whereas a botanist might think it means the recreational drug Catha edulis, and a zoologist would think it refers to the bird Bombycilla cedrorum. Such a mixup could result in confusion by a researcher studying human behaviour that now has data referring to flight activity or nest-building strategies. Moreover, the representation of each of these identifiers is incredibly varied, such as several different permutations of the same identifier being used to represent the human taxon. This is just a toy example of why these short “accession” style identifiers need to be unambiguously contextualized and actionable; this is most easily using the persistent web based identifier (http URI) instead of a simple database accession.
Even then, when referencing with links, you should be careful not to just copy the link you happen to find in the browser address bar, as that address may a) not be designed to persist, and — even if persistent — b) may nevertheless make it difficult for data integrators to know that you and someone else are referring to the same exact thing. Take for instance a cursory investigation of linked references to the SNCA gene record in NCBI; most references were just an accession (“gene:6622”) or symbol (“SNCA”) with no link. There were 38 such distinct short text representations. Links, when included at all, were represented one of twelve different ways:
- http://www.ncbi.nlm.nih.gov/gene/6622
- https://www.ncbi.nlm.nih.gov/gene/6622
- http://view.ncbi.nlm.nih.gov/gene/6622
- http://www.ncbi.nlm.nih.gov/gene?term=6622
- http://www.ncbi.nlm.nih.gov/gene/?term=6622
- http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Gene&term=6622
- http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=search&term=6622
- http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=gene&list_uids=6622
- http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=6469
- http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=retrieve&db=gene&list_uids=40966&dopt=full_report
- http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene&cmd=Retrieve&dopt=Graphics&list_uids=6622
- http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene&cmd=Retrieve&dopt=full_report&list_uids=6622
Even more permutations are bound to exist. The point is that a) we providers need to make referencing easier, and b) users should be aware of the links they use when referencing. Follow the provider’s documentation where available, or look up the durable representation at identifiers.org.
Data providers, take back the night!
This manuscript is also a guide to data providers: you can help by declaring how you want your identifiers referenced by others, whether in datasets or the literature, and where they should resolve to. Third party resolvers (such as N2T.net and identifiers.org) can be used as a clean and persistent layer that can redirect any identifier request to its final destination, for example,
http://n2t.net/ncbigene:6622 → http://www.ncbi.nlm.nih.gov/gene/6622 or
http://n2t.net/ZFIN:ZDB-FISH-150901-3034 → http://zfin.org/ZFIN:ZDB-FISH-150901-3034
A data provider can be more impactful and better utilized when databases, journals, and the Web at large can consistently and durably reference any given identified gene, organism, variant, etc. and link it to its authoritative source.
Authors, be inquisitive about ids!
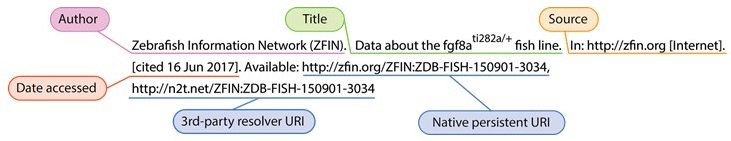
We and others have shown that model organisms, cell lines, antibodies, biospecimens, datasets, software, and other entities and resources are not uniquely referenced in the literature [2] hindering scientific reproducibility, attribution, and the ability to find knowledge. Biocurators spend a staggering amount of time chasing authors to uniquely identify which gene or genotype was referenced so as to associate other data – without which these data become lost to our collective knowledge. You can too – next time you use a model organism or reference a gene in a manuscript, consider referencing it using its authoritative, persistent, resolvable identifiers. More and more journals are supporting (and requiring) formal URI-based referencing of all kinds of entities. For example, an author might formally reference the fgf8ati282a/+ zebrafish line like so: ZDB-FISH-150901-3034 [3] (see also annotated version of this citation below), enabling readers to get directly back to the authoritative record in ZFIN, much in the same way they would for any other scholarly article. Formal bibliographic data citation (as opposed to mentions, or omission) not only facilitates reproducibility, it provides a more robust structure for credit mechanisms. References can also be formatted to include both the authoritative (native) link as well as the 3rd party meta-resolver. Link redundancy in references, especially when referenced by documents that can not (or should not) be updated can provide an important extra measure of protection against link rot.
Example bibliographic reference:
Take it home
A shared identifier system facilitates text mining for related content (e.g. genotype-phenotype data), resource tracking and reproducibility (e.g. which organism strain), content drift (e.g. gene function assessment over time), attribution, and dataset indexing. Proper identifier creation, maintenance, and referencing allows us to maintain the roads of bioinformatics. So reference the resources’ identifiers you use, don’t hesitate to ask for help, and report outages: keeping the roads free of potholes requires the distributed participation of all of us.
References
- Pearson H. Biology’s name game. Nature. 2001;411: 631–632. doi:10.1038/35079694
- Vasilevsky NA, Brush MH, Paddock H, Ponting L, Tripathy SJ, Larocca GM, et al. On the reproducibility of science: unique identification of research resources in the biomedical literature. PeerJ. 2013;1: e148. doi:10.7717/peerj.148
- Zebrafish Information Network (ZFIN). ZFIN Fish: fgf8ati282a/+. In: http://zfin.org [Internet]. [cited 16 Jun 2017]. Available: http://zfin.org/ZFIN:ZDB-FISH-150901-3034, http://n2t.net/ZFIN:ZDB-FISH-150901-3034
- McMurry JA, Juty N, Blomberg N, Burdett T, Conlin T, Conte N, et al. (2017) Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. PLoS Biol 15(6): e2001414. https://doi.org/10.1371/journal.pbio.2001414
Image credits:
- Photo of Dr. Ashburner is from https://doi.org/10.1371/journal.pcbi.1002081.g001
- Twitter screenshot is reproduced under fair use https://support.twitter.com/articles/20171959
- Other illustrations were done by Julie McMurry and Lilly Winfree



[…] In addition to the article, see also the PLOS Blog post Bad Identifiers are the Potholes of the Information Superhighway: Take-Home Lessons for Researchers. […]
[…] organism, to a gene, to a dataset, to a transcript, to a Gene Ontology term. A companion blog, “Bad Identifiers are the Potholes of the Information Superhighway”, aims to explain to the less nerdy what this means for the average researcher – consumers who, […]
[…] organism, to a gene, to a dataset, to a transcript, to a Gene Ontology term. A companion blog, “Bad Identifiers are the Potholes of the Information Superhighway”, aims to explain to the less nerdy what this means for the average researcher – consumers who, […]