Genetics and microbiome of cattle methane production: 2017 PLOS Genetics Research Prize Winning Research
The PLOS Genetics Editors-in-Chief and Senior Editors would like to congratulate: Rainer Roehe, Richard J. Dewhurst, Carol-Anne Duthie, John A. Rooke, Nest McKain, Dave W. Ross, Jimmy J. Hyslop, Anthony Waterhouse, Tom C. Freeman, Mick Watson and R. John Wallace, authors of the article chosen as the recipient of the 2017 PLOS Genetics Research Prize:
‘Bovine Host Genetic Variation Influences Rumen Microbial Methane Production with Best Selection Criterion for Low Methane Emitting and Efficiently Feed Converting Hosts Based on Metagenomic Gene Abundance’
In the winning research article [1], Rainer Roehe and colleagues from Scotland’s Rural College and the Universities of Edinburgh and Aberdeen set out to understand how the host genetics affects the rumen microbiome, and thus influence rumen microbial methane production and rumen microbial digestion.
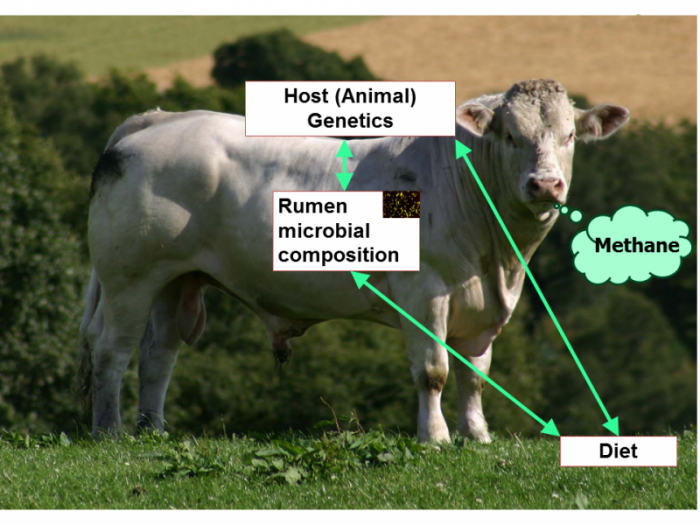
Anthropogenic production of methane contributes significantly to greenhouse gases and global warming, for which emissions from ruminant digestive tracts, e.g. gassy cows, are a major source. In their article, Rainer Roehe and colleagues tackle the genetics of cattle methane production with a metagenomic approach. This is a daunting research question: methane is produced by methanogenic archaea in the rumen of cattle, and therefore might be influenced by diet, host genetic factors, and the composition of the microbiome. The authors investigated the relative contributions of these factors by associating methane emissions measured individually in respiration chambers with genetic background (sire progeny groups) and diet. The analysis indicated a significant sire genetic effect on their progenies’ methane production and thus a contribution of the host genome. The authors also used the power of cattle family structures to identify that a fraction of the variation in the composition of the microbial community is controlled by the host genome. Roehe et al. further showed that the composition of the microbial community and particularly of the microbial genes is highly correlated with methane emissions. Importantly, the authors note that this metagenomic profile could provide a selection parameter that is much easier to implement in a real breeding program than direct measurement of methane emissions, which is difficult and expensive.
This study paves the way for an approach to reduce methane produced by cattle farming using modern breeding approaches. It should be noted that the study is necessarily based on a relatively small number of animals. The question of whether selection for a particular microbiome composition in cattle has any adverse effects, e.g. on the health of the animals, will need to be investigated. Nonetheless, the study by Roehe et al. advances our understanding of how genes and environment interact in the cattle gut, and provides a plausible scenario for an application that could greatly benefit society.
Corresponding author, Rainer Roehe says: “We are honored to receive this award from the high impact journal PLOS Genetics. Our research article answered the fundamental question that the host genetics shapes its own microbiome. In addition, the paper provides many new results, in particular based on the abundance of microbial genes to be used to identify mechanisms of the interactions between microbiome and host. In the future, we expect that the use of gastrointestinal microbial gene information will have a large impact in animal breeding, personalized medicine, nutritional recommendations, etc., in many different species.”
The PLOS Genetics Research Prize was born from the editors’ desire to recognize the outstanding work published in PLOS Genetics and launched in 2015 as part of the journal’s 10-year anniversary celebrations. Members of the genetics community nominated their favourite Research Article published in PLOS Genetics in 2016, and the Senior Editors selected the winning article from these nominations, based on scientific excellence and the community impact of the work.
Further details about the prize can be found here: https://www.staging.plos.org/genetics-research-prize/
Reference:
- Roehe R, Dewhurst RJ, Duthie C-A, Rooke JA, McKain N, Ross DW, et al. (2016) Bovine Host Genetic Variation Influences Rumen Microbial Methane Production with Best Selection Criterion for Low Methane Emitting and Efficiently Feed Converting Hosts Based on Metagenomic Gene Abundance. PLoS Genet 12(2): e1005846.
Featured image credit: December 2016 Issue Image. Immunofluorescent staining of 2-month-old ovarian sections. Image Credit: Meng-Wen Hu


[…] Genetics and microbiome of cattle methane production: 2017 PLOS Genetics Research Prize Winning Rese… PLoS Blogs (blog) […]
[…] «малометановый» крупный рогатый скот. Об этом сообщает сайт […]
[…] Genetics – Find out who won this year’s PLOS Genetics Research Prize and learn more about the winning research, submit a Topic Page, browse Up For a Challenge – Stimulating […]
[…] Genetics – Find out who won this year’s PLOS Genetics Research Prize and learn more about the winning research, submit a Topic Page, browse Up For a Challenge – Stimulating […]
[…] L’importance de ces productions naturelles de méthane est suffisante pour qu’un groupe de scientifiques suggère de modifier génétiquement les vaches pour qu’elles en produisent […]